Ans 1. The User friendly Comprehensive Antibiotic resistance Repository of Escherichia coli (u-CARE) is a manually curated database which dissects different strains of Escherichia coli on both reductive i.e. antibiotic reistance genes, SNP conferring antibiotic resitance, transcription factors involved in antibiotic resistance as well as holistic level i.e. genome specific Operons and pathogenics islands. ↑TOP
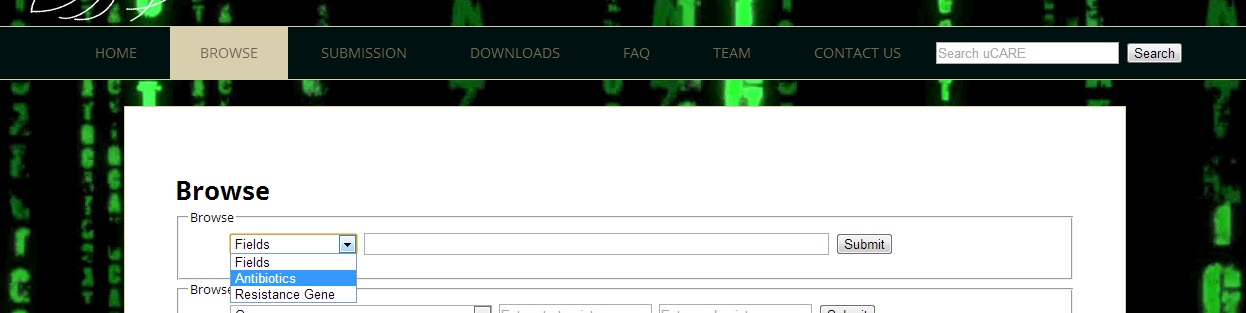
Ans 2.Antibiotics page can be retrieved by selecting "antibiotics" on the Browse through Fields tab and submitting antibiotic query. Antibiotic query can be Antibiotic name e.g. "ampicillin", pubchem cid e.g. "6249" or uCARE id. e.g. "UD0001". The list of all the antibiotics can also be found by selecting antibiotics from "BROWSE" drop down tab and submitting an empty form.
 ↑TOP
↑TOP
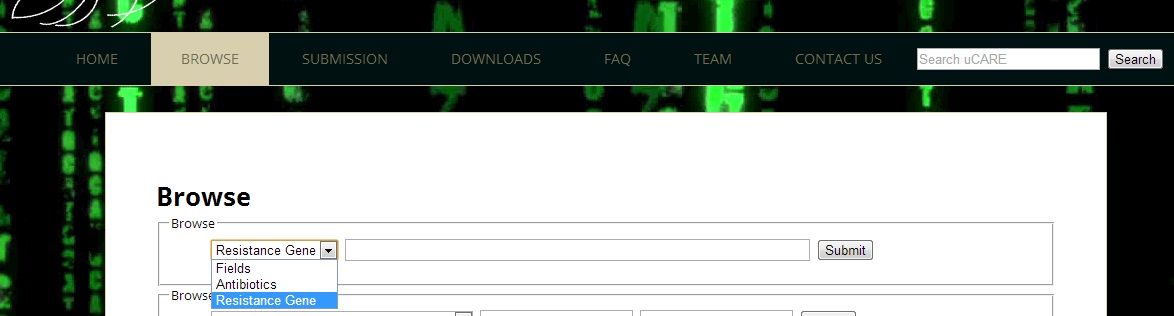
Ans 3.Resistance gene pages can be retrieved by selecting "Resistance gene" on the Browse through Fields tab and submitting the gene query. Gene query can be Gene name e.g. "sul1", NCBI gene id e.g. "12678136", uniprot id e.g. "G0XAY8" or genpept id e.g. "YP_006142104.1". The list of all resistance genes can also be found by selecting Resistance gene from "BROWSE" drop down tab and submitting an empty form.
 ↑TOP
↑TOP
Ans 4. The operons can be retrieved by selection specific E. coli genome from "Browse in OPERON" Genomes tab and submitting tha start and end range of E. coli genome.
 ↑TOP
↑TOP
Ans 5. The pathogenic islands can be retrieved by selection specific E. coli genome from "Browse in PAI" Genomes tab and submitting tha start and end range of E. coli genome.
 ↑TOP
↑TOP
Ans 6. uCARE's BLAST utility utilizes NCBI blastp and substitution matrix BLOSSUM62 with a expection value of 10.
NOTE:The BLAST utility works only for protein fasta sequence e.g.
>gi|386617477|ref|YP_006142104.1| dihydropteroate synthase protein Sul1 [Escherichia coli UMNK88]
MVTVFGILNLTEDSFFDESRRLDPAGAVTAAIEMLRVGSDVVDVGPAASHPDARPVSPADEIRRIAPLLD
ALSDQMHRVSIDSFQPETQRYALKRGVGYLNDIQGFPDPALYPDIAEADCRLVVMHSAQRDGIATRTGHL
RPEDALDEIVRFFEARVSALRRSGVAADRLILDPGMGFFLSPAPETSLHVLSNLQKLKSALGLPLLVSVS
RKSFLGATVGLPVKDLGPASLAAELHAIGNGADYVRTHAPGDLRSAITFSETLAKFRSRDARDRGLDHA
↑TOP
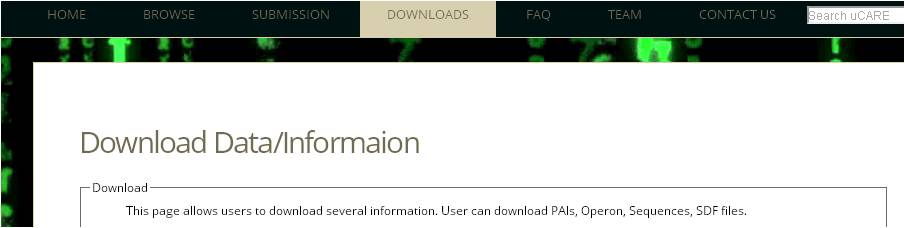
Ans 7. Complete pathogenic islands, Operons, resistance gene multifasta, and antibiotic sdf can be retrieved from "Download page".
 ↑TOP
↑TOP
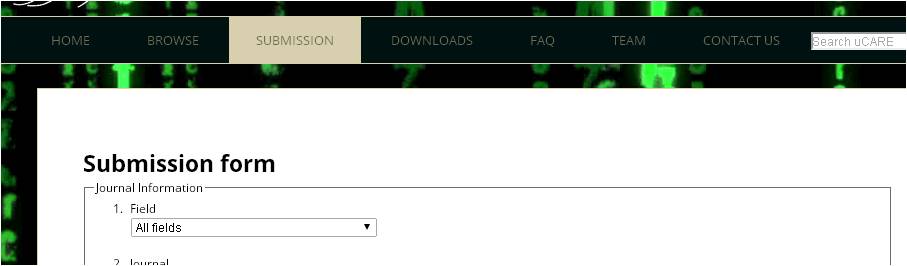
Ans 8. All the data submission can be done through "Submission page".
 NOTE: The database incorporates only already published data cited in pubmed or pubmed central.↑TOP
NOTE: The database incorporates only already published data cited in pubmed or pubmed central.↑TOP
Ans 9. Through this database, we envisage to build a platform where researchers working on Escherichia coli will be able to analyze E. coli pathogenesis on many dimensions. Apart from the gene and drug annotations provided, we plan to integrate primer BLAST for the genes and substiquently start a molecular structural prediction project to provide the structures of these canonical proteins involved in its drug resistance.